A core SCMB activity is catalyzing new collaborations at the math-bio interface through the funding of cross-disciplinary teams working on a research project of mutual interest. Far from being a collection of disparate topics, these projects organize into a coherent chain spanning temporal and spatial scales, each addressing a critical juncture of the genome-to-phenome pipeline. Each such "Seed Project" is co-lead by an interdisciplinary mathematician and a biosystems experimentalist, and includes two SCMB-supported junior researchers --- a biosystems graduate student and a mathematics postdoctoral scholar.
Critical to the success of these initiatives, the SCMB biosystems faculty have overlapping research interests ranging from molecular cell biology, quantitative genetics, and proteomics through single molecule microscopy all the way to developmental morphology and behavioral dynamics. Moreover, each brings substantial quantitative modeling and computational analysis expertise to the center. Dually, the mathematicians bring significant interdisciplinary experience, and also have many overlapping research interests. Of particular note are the foundational areas of algebra, topology, and geometry which have considerable untapped potential in biosystems applications. This is balanced by expertise in other areas, like combinatorics, probability and computation, where the interactions with biosystems are already better developed.
Research
-
Natasha Jonoska (USF) & Francesca Storici (GT): Discrete and topological models for DNA-RNA interactions
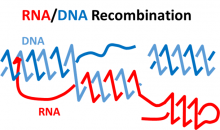
Genome stability is crucial for healthy life of cells and for transmission of accurate genetic information to progenies. Among the most dangerous DNA lesions are DNA double-strand breaks (DSBs), which slice both DNA strands. If not properly sealed, DSBs can cause mutations or chromosomal rearrangements leading to variety genetic disorders. RNA was recently discovered to be a direct template in DSB repair.
Read More -
Christine Heitsch (GT) & Annalise Paaby (GT): RNA structural ensembles in evolution
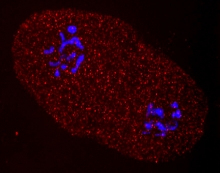
This project unites the mathematics of RNA folding with the biology of population evolution. The Paaby lab studies evolution and quantitative genetics in the nematode worm Caenorhabditis elegans and related Caenorhabditis species, including the role of molecular mechanisms in trait determination, while the Heitsch group focuses on the combinatorics of RNA base pairing, using different mathematical approaches to elucidate structure-function relationships for RNA sequences. Their SCMB Seed Project collaboration affords an unusual opportunity to rigorously quantify genomic fitness
Read More -
Julie Mitchell (ORNL) & Matt Torres (GT): Identifying disorder-to-order transitions in post-translationally modified proteins
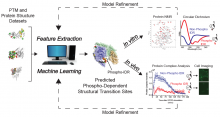
To better understand biological systems regulation at the molecular scale, Professors Mitchell and Torres will develop new predictive features and a machine learning model to identify regions within Instrinsically Disordered Regions (IDRs) able to undergo structural transitions following Post-Translational Modification (PTM). Our study of PTMs will initially focus on phosphorylation and use available protein structures for phosphorylated isoforms. We will pursue a machine learning approach based on evolutionary and sequence-based features.
Read More -
Scott McKinley (Tulane) & Christine Payne (Duke): Stochastic modeling in cellular internalization and transport
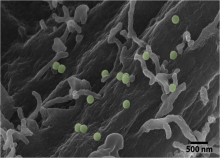
Continuing advances in live cell imaging and single particle tracking techniques have led to remarkable new insights for cellular biologists, while presenting profound challenges for probabilists and statisticians. A wide array of biological agents including organelles, viruses, and cellular nutrients move through crowded media in complex geometries. These interactions bring forth stunningly organized emergent behavior that form the basis of cellular health and disease.
Read More -
Elena Dimitrova (Clemson) & Melissa Kemp (GT): Modeling emergent patterning within pluripotent colonies through Boolean canalizing functions
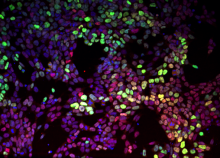
Profs. Melissa Kemp and Elena Dimitrova will investigate how stem cells pattern within colonies due to specific cell-cell communication. Multicellular populations give rise to emergent features such as patterns based upon the collective communication between neighboring and distant cells. Transitions in early development from pluripotent systems to differentiated, structurally-ordered tissue provide an ideal testbed to study generalizable properties of cellular coordination. Prof.
Read More -
Peter Bubenik (Florida) & Hang Lu (GT): Topological data analysis to understand genetic control of morphological phenotype
This new collaboration will use topological data analysis to summarize morphological changes in order to understand the genetic basis of phenotype. Hang Lu has developed a series of microfluidic devices and automation tools that can consistently generate thousands of stacks of images of fluorescent markers in individual nematodes at diffraction-limited spatial resolution. Peter Bubenik has developed topological tools for summarizing the geometry of complex multiscale biological data and analyzing these summaries using statistics and machine learning. The Lu lab will generate large collections of images from C. elegans carrying morphological markers. The Bubenik group will apply their tools to analyze the data from the Lu lab.
Read More -
Greg Blekherman (GT) & Dan Goldman (GT): Optimization of limbless locomotion via algebraic kinematics

Discovering principles of locomotion in complex terrain remains a challenge: how do organisms coordinate their enormous numbers of degrees of freedom to generate changes in body shape which lead to emergent robustness, stability and efficiency? Of interest to us in this project is limbless locomotion: snakes must coordinate the interaction of their flexible trunks with environmental heterogeneities (rocks, branches) to generate propulsion.
Read More


