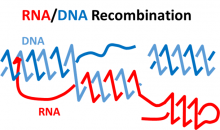
Genome stability is crucial for healthy life of cells and for transmission of accurate genetic information to progenies. Among the most dangerous DNA lesions are DNA double-strand breaks (DSBs), which slice both DNA strands. If not properly sealed, DSBs can cause mutations or chromosomal rearrangements leading to variety genetic disorders. RNA was recently discovered to be a direct template in DSB repair. Although in recent years it has become more evident that RNA-DNA hybrids play essential roles in cell functions, there are almost no mathematical studies on the molecular complex and the circumstances of its emergence. Senior personnel Jonoska (USF, Math) and Storici (Georgia Tech, Biological Sciences) have an established, independent history of studying RNA-guided DNA modifications and in this project they start a collaboration with a goal to develop topological and graph theoretical methods to study the basic relationship between RNA and DNA in the context of genome stability and modification. The studies combine molecular biology experiments and mathematical models enhancing both disciplines. Using both, the molecular structure and sequence analysis, we seek to understand the best RNA-DNA conformation that would guide the DNA repair. The experiments help to build the structural description of the RNA-DNA hybrids, while the developed models drive the design of the tests in vivo and make predictions of DNA repair outcomes, which, in turn, can be validated in vivo.


