Continuing advances in live cell imaging and single particle tracking techniques have led to remarkable new insights for cellular biologists, while presenting profound challenges for probabilists and statisticians. A wide array of biological agents including organelles, viruses, and cellular nutrients move through crowded media in complex geometries. These interactions bring forth stunningly organized emergent behavior that form the basis of cellular health and disease. Every component of intracellular movement is intrinsically random and, formerly, the dominant mathematical question was: "How have these agents evolved so that they can overcome stochasticity and be robust?" Now that we can directly observe spatial organization and the mechanisms supporting cell signaling, we recognize that the real question is: "How have these agents evolved to exploit stochasticity in order to be so robust?" The Payne-McKinley collaboration will focus on endocytosis, the process by which cells bind, internalize, and transport extracellular cargo. We will collect and use single particle tracking data to study how natural diffusion processes, coupled with biochemical interactions and ATP-dependent active transport mechanisms, distribute material to necessary locations on the cell surface and within cells. Moreover, we will seek to understand how subtle rules for transport and search can optimize delivery of cargo and organization of biomaterials.
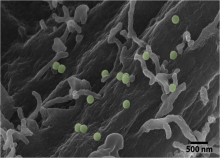
Cells interact with nutrients, viruses, and toxins through endocytosis, the cellular internalization and transport of bioparticles. The initial internalization step requires specific cell surface receptors to access the cell. However, initial contact with the cell surface is unlikely to occur at a dedicated receptor, but rather a random location on the cell surface. The bioparticle must then sample the cell surface to bind to the correct receptor. Using fluorescently-labeled, functionalized, nanoparticles (NPs) as a model system, we will use single particle tracking to study how bioparticles bind and unbind from the cell surface prior to binding to a dedicated cell surface receptor. We will use Bayesian techniques to select from a number of possible mathematical models to understand how cellular binding affinity affects the time it takes for bioparticles to find their dedicated cell surface receptors and the systems-level benefit in sampling of the plasma membrane space.


